By Carol Clark
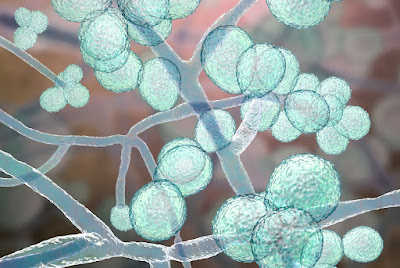
A novel technique to test platelet function within a person’s blood sample is faster, easier and more precise than methods currently in use, an experimental study shows.Tuesday, December 12, 2023
New tool to analyze blood platelets holds major medical potential
Tuesday, December 5, 2023
Building boom boosts malaria-carrying, invasive mosquito in Ethiopia
Monday, November 20, 2023
Birds set foot near South Pole in Early Cretaceous, Australian tracks show
The discovery of 27 avian footprints on the southern Australia coast — dating back to the Early Cretaceous when Australia was still connected to Antarctica — opens another window onto early avian evolution and possible migratory behavior.
PLOS ONE published the discovery of some of the oldest, positively identified bird tracks in the Southern Hemisphere, dated to between 120 million and 128 million years ago.
“Most of the bird tracks and body fossils dating as far back as the Early Cretaceous are from the Northern Hemisphere, particularly from Asia,” says Anthony Martin, first author of the study and a professor in Emory University’s Department of Environmental Sciences. “Our discovery shows that there were many birds, and a variety of them, near the South Pole about 125 million years ago.”
Related:
Tell-toe toes point to oldest-known bird tracks from Australia
Paleontologist explores a billion years of animals breaking up rocks, bones, shells and wood
Friday, November 10, 2023
NSF funds holistic approach to help farmers adapt to climate change
By Carol Clark
The National Science Foundation (NSF) awarded Emily Burchfield, Emory assistant professor of environmental sciences, $1.6 million to lead efforts to identify emerging pressures on agriculture in Georgia, Iowa and Ohio and to develop predictive models to help farmers and policymakers weather these changes.
“In a nutshell, we’re trying to understand what climate change will mean for agriculture in these three states,” Burchfield says. “We’ll be integrating biophysical projections based on environmental data with insights gathered from farmers and agricultural experts.”
The goal is to develop possible scenarios for the impacts of climate change — along with the evolving technical, socioeconomic and political landscapes in each state — for how and where crops could be grown over the next 30 to 40 years. The researchers will create a public, online tool to allow farmers and policymakers to explore the possible futures of agriculture at regional and state levels and to support their efforts to manage these scenarios.
The grant is part of the NSF Dynamics of Integrated Socio-Environmental Systems Program (DISES).
“Traditionally, the NSF has mainly split programs into the social sciences and the natural sciences but DISES is one of their newer programs that joins the two, looking at how nature affects people and people affect nature,” Burchfield says. “Coupling human and natural systems in theoretical frameworks allows us to take on some of the grand challenges that we’re facing, like climate change and food and water security.”
Burchfield is principal investigator for the project, which also includes researchers from Arizona State University, Ohio State University and the University of Nebraska, Lincoln.
A range of agricultural systems
While the two main crops in both Iowa and Ohio are corn and soy, agriculture in Georgia is far more diverse. The state leads the nation in the production of peanuts, pecans, blueberries and spring onions and is also a leading producer of cotton, watermelon, peaches, cucumbers, sweet corn, bell peppers, tomatoes, cantaloupes, rye and cabbage.
Agriculture contributes nearly $70 billion annually to Georgia’s economy and one in seven Georgians works in agriculture, forestry or related fields, according to the Georgia Farm Bureau.
“Compared to other parts of the country, Georgia is incredibly diverse not just in terms of what is grown in the state but in terms of who grows it,” Burchfield says. “A lot of exciting changes are happening in the state — citrus production is moving into South Georgia. And the biggest organic farm east of the Mississippi is located in Georgia, producing carrots.”
While California currently produces the bulk of the nation’s produce, that state is facing significant challenges for water availability, Burchfield notes. “Georgia has a unique opportunity to expand its fresh-produce production to help meet future demand,” she says. “We want to provide farmers the resources they need to capitalize on such trends.”
Building tools for the future of farming
Burchfield’s research combines spatial-temporal social and environmental data to understand the future of food security in the United States, including the consequences of a changing climate.
For the current project, the researchers will draw from available climate, soil and land-use data to create biophysical models for how changes in climate will affect where and how particular crops can be grown. These models will be integrated with data gathered from surveys and focus groups conducted with agricultural experts, climatologists and farmers working the land throughout Georgia, Iowa and Ohio.
The project aims to get input from a diverse range of farmers growing different crops and using different management practices.
“There is already a lot of work on what climate change may mean for agriculture in general,” Burchfield says. “But what climate change means for an individual farm must be filtered through issues particular to that farm. So many dimensions that matter deeply to farmers are not included in policy discussions about agriculture.”
Farmers will be asked what information and resources they need to sustain their operations and to adapt to climate change. “We want to understand the vision that farmers have for the future of their farms,” Burchfield says. “What would they would like to see happen? What do they see as the barriers and bridges to achieving that vision?”
The public, online tool that the researchers develop will include interactive maps for crop forecasts by region. It will also provide information to guide policymakers and to help farmers adapt to the changes ahead.
“It’s impossible to accurately say exactly what’s going to happen in the future,” Burchfield says. “But combining biophysical data with an understanding of the technical, economic and political changes emerging in each of these states, along with the expertise of our farmers, will allow us to forecast trends for how suitable particular regions will be for growing certain crops. The bottom line is we are pulling together the best information available to give a sense of the emergent opportunities in the state for agriculture as well as the emergent challenges.”
Related:
Emory breaking new ground for climate-smart agriculture
Climate change on course to hit U.S. Corn Belt especially hard
Thursday, November 9, 2023
New antimicrobial shuts down bacterial growth without harming human cells
By Carol Clark
Scientists have shown how a molecule with broad-spectrum antibiotic activity works by disabling a process vital to bacterial growth without affecting the normal functioning of human cells. mBio, a journal of the American Society for Microbiology, published the work, led by researchers at Emory University and Pennsylvania State University.
The molecule, known as KKL-55, is one of a suite of recently identified molecules that interfere with a key bacterial mechanism known as trans-translation, essentially shutting down the ability of bacteria to grow.
“We’re opening a promising pathway for the development of new antibiotics to treat drug-resistant infections,” says Christine Dunham, co-corresponding author of the paper and a professor in Emory’s Department of Chemistry and the Emory Antibiotic Resistance Center.
Kenneth Keiler, a professor in the Department of Biochemistry and Molecular Biology at Pennsylvania State, is co-corresponding author of the paper.
First authors are Ha An Nguyen, who did the work as an Emory chemistry PhD candidate and has since graduated and works at Memorial Sloan Kettering, and Neeraja Marathe, a graduate student at Pennsylvania State.
A growing global threat
Antimicrobial-resistant infections have long been a public health threat. The situation grew even worse during the COVID-19 pandemic with increased antibiotic use and less prevention actions, according to the U.S. Centers for Disease Control and Prevention (CDC).
The CDC estimates that at least 2.8 million antimicrobial-resistant infections continue to occur in the United States each year, killing more than 35,000 people. Globally, the World Health Organization projects that these infections will cause up to 10 million deaths annually by 2050 if new antibiotics are not developed.
While antibiotics can save lives, any time they are used they can also contribute to the problem of resistance. Bacteria keep evolving new weapons as a defense against drugs, even as scientists work on developing new strategies to disarm bacteria.
Cross-toxicity, or harmful effects on humans, is another key drawback of some of the drugs used in a last-ditch effort to kill antibiotic-resistant bacteria.
Avoiding cross-toxicity
Dunham and Keiler are avoiding the problem of cross-toxicity by focusing on the inhibition of a mechanism unique to bacteria — trans-translation. This mechanism is vital to the proper functioning of the bacterial ribosome.
Keiler, a molecular geneticist and biochemist, first identified trans-translation in bacteria and is an expert in how it functions. Dunham, a structural biologist, is an expert in the human ribosome. She uses advanced biochemistry and structural biology techniques to understand the mechanics of molecular interactions.
“Our individual areas of expertise mesh well for this project,” Dunham says. “By collaborating, we are able to take the science further, faster.”
A cellular protein factory
The ribosome is an elaborate macromolecular machine within a cell that operates like a factory to manufacture proteins. Proteins are the machines that make cells run while nucleic acids such as DNA and RNA store the blueprints for life. The ribosome is made mostly of RNA, which does not just store information but can also act as an enzyme, catalyzing chemical reactions.
In a human cell, messenger RNA (mRNA), containing the instructions for manufacturing a protein, originates in the nucleus. While still in the nucleus, mRNA undergoes an elaborate quality-control process. It must pass inspection before getting exported to translate the information it contains into a protein.
“A lot of mRNAs have defects,” Dunham says. “Human cells have efficient ways to test mRNAs and ultimately remove the defective ones.”
Bacterial cells, however, have no nucleus or organized center for quality control.
“Bacteria wants to grow, grow and grow, which requires the ribosome to make a lot of proteins,” Dunham says. “But when mRNA has defects, there is little to no quality control. When the ribosome encounters a defective mRNA protein, synthesis gets stalled.”
The trans-translation process “rescues” ribosomes stalled due to such defects, in order to maintain proper protein synthesis and cell viability in bacteria.
How KKL-55 works
Using a high-throughput screening process, the Keiler lab has identified dozens of molecules that inhibit trans-translation in bacteria.
For the current paper, the researchers focused on understanding how one of these molecules, KKL-55, performs this trick. They used the high-powered structural biology technique of X-ray crystallography to capture KKL-55 in action as it interacted with a protein required for translation.
The results showed how KKL-55 blocks trans-translation by binding to elongation factor thermos-unstable (EF-Tu). EF-Tu is a protein that interacts with transfer RNA molecules, which play a key role in protein synthesis, and also transfer-messenger RNA, an RNA molecule required for the trans-translation pathway.
“We got lucky,” Dunham says. “There are dozens of steps involved in the process that KKL-55 could have inhibited and we might have had to test for each one. But the results are clear-cut. It shuts down trans-translation right at the beginning by preventing EF-Tu from binding to tmRNA.”
Determining the mechanism by which a molecule works to inhibit bacteria is a critical step toward developing a new antibiotic for clinical use. The next step is to test the efficacy of KKL-55 to treat a bacterial infection in a mouse model.
In 2021, the research team published their finding that a group of trans-translation inhibitors known as acylaminooxadiazoles clear multiple-drug-resistant Neisseria gonorrhoeae infection in mice after a single oral dose. That work is now advancing to clinical trials.
Dozens more trans-translation inhibitors await the team’s investigation. Each represents a potential new weapon to help humans stay on top in the arms race with drug-resistant bacteria.
Co-authors of the current paper include Alexandra Nagy (a former National Institutes of Health FIRST Institutional Research and Academic Career Development postdoctoral fellow at Emory who is now at Earlham College); as well as John Alumasa and Michael Vazquez (both from Pennsylvania State University). The work was funded by the National Institutes of Health, the National Institute of General Medical Sciences.
Related:
Images of enzymes in action reveal secrets of antibiotic-resistant bacteria
Monday, October 23, 2023
Emory breaking new ground for climate-smart agriculture in the Southeast
Three Emory University researchers received $5,100,000 as part of a United States Department of Agriculture (USDA) project to help measure and promote climate-smart practices that support small-scale, diversified vegetable farmers in the Southern Piedmont. A plateau below the Appalachian Mountains and above the coastal plain, the Southern Piedmont is a banana-shaped region spanning a bit of eastern Alabama, up across part of northern Georgia and into North and South Carolina and Virginia.
Emory is one of 12 organizations involved in the $25 million project, headed by the Rodale Institute and titled “Quantifying the Potential to Reduce Greenhouse Gas Emissions and Increase Carbon Sequestration by Growing and Marketing Climate-Smart Commodities in the Southern Piedmont.”
The five-year project is part of the USDA’s Partnerships for Climate-Smart Commodities initiative. “This effort will increase the competitive advantage of U.S. agriculture both domestically and internationally, build wealth that stays in rural communities and support a diverse range of producers and operation types,” USDA Secretary Tom Vilsack says of the initiative.
The Emory team encompasses three faculty from the Department of Environmental Sciences: Emily Burchfield, Eri Saikawa and Debjani Sihi.
• Burchfield combines spatial-temporal social and environmental data to understand the future of food security in the United States.
• Saikawa is an atmospheric chemist who models global soil nitrous oxide emissions and quantifies soil greenhouse gas fluxes.
• Sihi is an environmental biogeochemist who researches soil organic matter dynamics and greenhouse gas emissions from natural and managed systems.
Related:
Climate change on course to hit U.S. corn belt especially hard
Diverse land cover boosts yields for major U.S. crops, study finds
Soil quality critical to help some U.S. crops weather climate change
Thursday, October 19, 2023
Math trio makes new points about size of the smallest triangle
Wednesday, September 20, 2023
Analyzing ways to help golden eagle populations weather wind-energy growth
By Carol Clark
Wind energy is a major component of the U.S. clean-energy goals. Already one of the fastest growing and lowest-cost sources of electricity in the country, it is poised for even more rapid growth, according to the U.S. Department of Energy.
Wind power, however, does not come without tradeoffs, including some negative impacts on wildlife. Throughout the United States, for example, it’s been estimated that as many as three golden eagles per wind farm are killed each year by wind turbines.
“Renewable energy sources, including wind energy, are critical for us to achieve a net-zero emissions future,” says Eric Lonsdorf, assistant professor of environmental sciences at Emory University. “We need to address conflicts between renewable energy and wildlife conservation so that we can combat climate change while also limiting damage to biodiversity.”
Lonsdorf and colleagues are developing data-driven methods to determine how much effort is needed to save golden eagles in order to offset the impact of wind turbines on their populations.
The Journal of Wildlife Management recently published their latest model for calculating the benefit of one mitigation strategy — removal of large, road-killed animals that can lead to golden eagles getting hit by cars.
Quantifying the benefits of natural capital
Lonsdorf is an expert in natural capital, or the quantifiable benefits that nature provides humans. He translates ecological principles and data into computer models that enable industry leaders and policymakers to better manage natural resources.
Co-authors of the current study include James Gerber and Deepak Ray, from the University of Minnesota; Steven Slater, from HawkWatch International; and Taber Allison, from the Renewable Energy Wildlife Institute.
The U.S. Fish and Wildlife Service (FWS) monitors golden eagle populations, which are protected through the Bald and Golden Eagle Protection Act and the Migratory Bird Treaty Act. Threats to golden eagles include loss of habitat and prey.
Additional threats that are directly linked to human activities include illegal shootings, electrocution at power poles, lead poisoning from consuming parts of bullets in the entrails of deer carcasses discarded at the site of hunters’ kills, collisions with cars at sites where the birds are scavenging roadkill and collisions with the blades of a wind turbine.
Across the western United States, hundreds of wind turbines have gone up in sage-brush flats that are part of golden eagles’ core habitat, and many more turbines are planned. In order to meet the permit requirements of the FWS, wind-energy companies must agree to mitigate their impact on the animals by offsetting the predicted number of golden eagles that will fly into their turbines each year.
Currently, the only offset strategy approved by the FWS for wind-energy companies is to retrofit power poles to prevent golden eagles from becoming electrocuted.
Adding empirical data
For the past five years, Lonsdorf and his colleagues have combined their expertise to develop a range of potential offset strategies for golden eagle fatalities.
Their current paper — an updated model for golden eagle mortality due to vehicle collisions based on data from Wyoming — considered myriad factors such as the population density for golden eagles in the region, the number and size of deer roadkill carcasses expected and the traffic volume on the roads. The model also incorporated observational evidence of eagle-carcass roadside interactions obtained by motion-triggered cameras, data that was lacking in a previous model the researchers created.
The addition of this empirical data allowed the researchers to make estimates for how long a golden eagle typically spends at a carcass, how the decay rate of the carcass affects the number of visits from eagles and the effects of seasonality on the scavenging behavior of the eagles.
The model results suggest that carcass relocation is a viable golden eagle mitigation strategy that could save up to seven golden eagles annually in some Wyoming counties. On average, the model indicates that the prompt removal of four roadside carcasses would save at least one golden eagle.
The researchers can make a user-friendly version of the prediction framework available to the FWS and wind-energy companies if the FWS decides to approve carcass removal as an eagle mortality offset strategy.
“We’re taking basic information about golden eagle ecology in the Anthropocene and developing it into predictive frameworks for how to protect them,” Lonsdorf says. “As wind energy continues to grow, more mitigation strategies will likely be needed. Our goal is to provide scientific evidence for a portfolio of methods to help accomplish a zero-net loss of golden eagles from wind-energy facilities.”
Related:
Valuing 'natural capital' vital to avoid next pandemic, global experts warn
International trade bans on endangered species tend to help mammals but hurt reptiles
Wednesday, September 13, 2023
Natural compound found in plants inhibits deadly fungi
By Carol Clark
A new study finds that a natural compound found in many plants inhibits the growth of drug-resistant Candida fungi — including its most virulent species, Candida auris, an emerging global health threat. The journal ACS Infectious Diseases published the discovery led by scientists at Emory University.
Laboratory-dish experiments showed that the natural compound, a water-soluble tannin known as PGG, blocks 90% of the growth in four different species of Candida fungi. The researchers also discovered how PGG inhibits the growth: It grabs up iron molecules, essentially starving the fungi of an essential nutrient. By starving the fungi rather than attacking it, the PGG mechanism does not promote the development of further drug resistance, unlike existing antifungal medications.
Laboratory-dish experiments also showed minimal toxicity of PGG to human cells.
“Drug-resistant fungal infections are a growing healthcare problem but there are few new antifungals in the drug-development pipeline,” says Cassandra Quave, senior author of the study and associate professor in Emory School of Medicine’s Department of Dermatology and the Center for the Study of Human Health. “Our findings open a new potential approach to deal with these infections, including those caused by deadly Candida auris.”
C. auris is often multidrug-resistant and has a high mortality rate, leading the Centers for Disease Control and Prevention (CDC) to label it a serious global health threat.
“It’s a really bad bug,” says Lewis Marquez, first author of the study and a graduate student in Emory’s molecular systems and pharmacology program. “Between 30 to 60% of the people who get infected with C. auris end up dying.”
An emerging threat
Candida is a yeast often found on the skin and in the digestive tract of healthy people. Some species, such as Candida albicans, occasionally grow out of control and cause mild infections in people. In more serious cases, Candida can invade deep into the body and cause infections in the bloodstream or organs such as the kidney, heart or brain.
Immunocompromised people, including many hospital patients, are most at risk for invasive Candida infections, which are rapidly evolving drug resistance.
In 2007, the new Candida species, C. auris, emerged in a hospital patient in Japan. Since then, C. auris has caused health care-associated outbreaks in more than a dozen countries around the world with more than 3,000 clinical cases reported in the United States alone.
A ‘natural’ approach to drug discovery
Quave is an ethnobotanist, studying how traditional people have used plants for medicine to search for promising new candidates for modern-day drugs. Her lab curates the Quave Natural Product Library, which contains 2,500 botanical and fungal natural products extracted from 750 species collected at sites around the world.
“We’re not taking a random approach to identify potential new antimicrobials,” Quave says. “Focusing on plants used in traditional medicines allows us to hone in quickly on bioactive molecules.”
Previously, the Quave lab had found that the berries of the Brazilian peppertree, a plant used by traditional healers in the Amazon for centuries to treat skin infections and some other ailments, contains a flavone-rich compound that disarms drug-resistant staph bacteria. Screens by the Quave lab had also found that the leaves of the Brazilian peppertree contain PGG, a compound that has shown antibacterial, anticancer and antiviral activities in previous research.
A 2020 study by the Quave lab, for instance, found that PGG inhibited growth of Carbapenem-resistant Acinetobacter baumannii, a bacterium that infects humans and is categorized as one of five urgent threats by the CDC.
The Brazilian peppertree, an invasive weed in Florida, is a member of the poison ivy family. “PGG has popped up repeatedly in our laboratory screens of plant compounds from members of this plant family,” Quave says. “It makes sense that these plants, which thrive in really wet environments, would contain molecules to fight a range of pathogens.”
Experimental results
The Quave lab decided to test whether PGG would show antifungal activity against Candida. Laboratory-dish experiments demonstrated that PGG blocked around 90% of the growth in 12 strains from four species of Candida: C. albicans, multidrug-resistant C. auris and two other multidrug-resistant non-albicans Candida species.
PGG is a large molecule known for its iron-binding properties. The researchers tested the role of this characteristic in the antifungal activity.
“Each PGG molecule can bind up to five iron molecules,” Marquez explains. “When we added more iron to a dish, beyond the sequestering capacity of the PGG molecules, the fungi once again grew normally.”
Dish experiments also showed that PGG was well-tolerated by human kidney, liver and epithelial cells. “Iron in human cells is generally not free iron,” Marquez says. “It is usually bound to a protein or is sequestered inside enzymes.”
A potential topical treatment
Previous animal studies on PGG have found that the molecule is metabolized quickly and removed from the body. Instead of an internal therapy, the researchers are investigating its potential efficacy as a topical antifungal.
“If a Candida infection breaks out on the skin of a patient where a catheter or other medical instrument is implanted, a topical antifungal might prevent the infection from spreading and entering into the body,” Marquez says.
As a next step, the researchers will test PGG as a topical treatment for fungal skin infections in mice.
Meanwhile, Quave and Marquez have applied for a provisional patent for the use of PGG for the mitigation of fungal infections.
“These are still early days in the research, but another idea that we’re interested in pursuing is the potential use of PGG as a broad-spectrum microbial,” Quave says. “Many infections from acute injuries, such as battlefield wounds, tend to be polymicrobial so PGG could perhaps make a useful topical treatment in these cases.”
Scientists from the University of Toronto are co-authors of the paper, including Yunjin Lee, Dustin Duncan, Luke Whitesell and Leah Cowen. Whitesell and Cowen are co-founders and shareholders in Bright Angel Therapeutics, a platform company for development of antifungal therapeutics, and Cowen is a science advisor for Kapoose Creek, a company that harnesses the therapeutic potential of fungi.
The work was supported by grants from the National Institutes of Health, National Center for Complementary and Integrative Health; the Jones Center at Ichauway, the CIHR Frederick Banting and Charles Best Canada Graduate Scholarship and the Canadian Institutes of Health Research Foundation.
Related
Extracts from two wild plants inhibit COVID virus, study finds
Scientists identify chemicals in noxious weed that 'disarm' deadly bacteria
Friday, September 8, 2023
NIH funds Emory center to advance cellular mechanics
By Carol Clark
The National Institutes of Health (NIH) awarded Emory University $5.6 million to establish a national center to advance pioneering technology for cellular mechanics. The center is directed by Khalid Salaita, Emory professor of chemistry, whose lab developed the first sensors for detecting cell-receptor forces at the molecular level.
“We’ve been working on our molecular-force probes for more than a decade,” Salaita says. “We’ve demonstrated that these probes can be used to visualize, measure and map cellular forces down to the level of piconewtons. The center allows us to get this technology into the hands of end users — researchers in the biomedical sciences.”
The Center for Molecular Mechanobiology encompasses labs from seven leading research institutions including: Children’s Hospital of Philadelphia, Dana-Farber Cancer Institute, Emory, Georgia Tech, Memorial Sloan Kettering, University of Utah and Vanderbilt University.
The center members will use the molecular-force probes to investigate the biomechanics of processes such as the clotting of blood cells, the response of immune cells to an infection and the migration of cancer cells. Better understanding these processes may lead to the development of new treatments and therapies for a range of diseases and disorders.
In addition to supplying the technology, the center will train researchers to use the molecular-force probes and help adapt the technology to answer specific biomedical research questions.
“Working directly with the research community will help us to further refine and optimize the technology,” Salaita says. “We envision that measuring cellular forces will soon become part of the standard repertoire of biochemical techniques that scientists use to study living systems.”
The center’s associate directors are Yonggan Ke (associate professor in the Wallace H. Coulter Department of Biomedical Engineering at Emory and Georgia Tech) and Alexa Mattheyses (associate professor in the Department of Cell Developmental and Integrative Biology at the University of Alabama).
The five-year award from the National Institute of General Medical Sciences is part of the NIH Biomedical Technology Optimization and Dissemination Centers program. The goal is to optimize and disseminate state-of-the-art, late-stage biomedical technologies.
The first detailed view of mechanical forces
The Salaita lab works at the intersection of chemistry, biology and the physical sciences. It uses the building blocks of nature — nucleic acids — to create synthetic micro motors and probes for investigating fundamental questions of biology.
The molecular-force probes, developed by the Salaita lab in 2011, provide the first detailed view of the mechanical forces on the surface of a cell. The technology can detect mechanical forces as fleeting as the blink of an eye and as faint as piconewtons — about one billionth the weight of a paperclip.
The probes are made from strands of synthetic DNA tagged with fluorescence so that they function like molecular beacons, shining when they sense force. The technique is noninvasive, does not modify the cell and can be done with a standard fluorescence microscope.
In 2014, the lab used the new method to demonstrate how adherent cells — the kind that form the architecture of all multicellular organisms — mechanically sense their environments, migrate and stick to things.
In 2016, the molecular-force probes provided the first direct evidence for the mechanical forces of T cells, the security guards of the immune system. The lab’s experiments on T cells drawn from mice showed how they use a kind of mechanical “handshake” to test whether a cell they encounter is a friend or a foe.
In 2017, the lab shined its molecular beacons on platelets, the cells in the blood whose job is to stop bleeding by sticking together to form clots and plug up a wound. That work revealed the key molecular forces on platelets that trigger the clotting process.
In 2020, the lab and its collaborators combined advances in optical imaging with the molecular-force probes to capture forces at a resolution of 25 nanometers — far shorter than the length of a light wave. “That resolution is akin to being on the moon and seeing the ripples caused by raindrops hitting the surface of a lake on the Earth,” Salaita said at the time.
Key technological goals
The Center for Molecular Mechanobiology will build on this foundational work of the Salaita lab. It will focus on three key technological development goals:
• Optimizing the highest-resolution technique of the molecular-force probes so that it can be applied to a range of research questions.
• Tagging cells based on their force level in order to use force as a marker to barcode cells and their receptors. The idea is to classify the mechanics of individual cells and then link these classifications to gene-expression levels to study the cause-and-effect relationships.
• Amplifying the molecular-force signals to better understand the role of even the weakest forces involved in cellular mechanics, including those involved in the immune response.
Researchers from throughout the country will come to the Center for Molecular Mechanobiology to receive hands-on training in the molecular-force probes and then return to their home labs to become ambassadors for the technology.
“We’ll be adding a whole other layer of information for researchers working on everything from designing vaccines to cancer immunotherapy agents,” Salaita says.
Decades ago, he points out, complicated techniques such as crystallography, PCR and mass spectrometry were not frequently used but have since become routine workhorses in the biomedical sciences.
“We are catalyzing the process of spreading our technology so that studying biomechanics also becomes common and routine in biology,” Salaita says. “Molecular forces are a missing piece to understanding the way biology works.”
Related:
‘Firefly’ imaging method makes cellular forces visible
Chemists reveal the force within you
T cells use ‘handshakes’ to sort friends from foes
New methods reveal the mechanics of blood clotting
@font-face
{font-family:"Cambria Math";
panose-1:2 4 5 3 5 4 6 3 2 4;
mso-font-charset:0;
mso-generic-font-family:roman;
mso-font-pitch:variable;
mso-font-signature:-536870145 1107305727 0 0 415 0;}@font-face
{font-family:Calibri;
panose-1:2 15 5 2 2 2 4 3 2 4;
mso-font-charset:0;
mso-generic-font-family:swiss;
mso-font-pitch:variable;
mso-font-signature:-536859905 -1073732485 9 0 511 0;}p.MsoNormal, li.MsoNormal, div.MsoNormal
{mso-style-unhide:no;
mso-style-qformat:yes;
mso-style-parent:"";
margin:0in;
mso-pagination:widow-orphan;
font-size:12.0pt;
font-family:"Calibri",sans-serif;
mso-ascii-font-family:Calibri;
mso-ascii-theme-font:minor-latin;
mso-fareast-font-family:Calibri;
mso-fareast-theme-font:minor-latin;
mso-hansi-font-family:Calibri;
mso-hansi-theme-font:minor-latin;
mso-bidi-font-family:"Times New Roman";
mso-bidi-theme-font:minor-bidi;}a:link, span.MsoHyperlink
{mso-style-priority:99;
color:#0563C1;
mso-themecolor:hyperlink;
text-decoration:underline;
text-underline:single;}a:visited, span.MsoHyperlinkFollowed
{mso-style-noshow:yes;
mso-style-priority:99;
color:#954F72;
mso-themecolor:followedhyperlink;
text-decoration:underline;
text-underline:single;}.MsoChpDefault
{mso-style-type:export-only;
mso-default-props:yes;
font-family:"Calibri",sans-serif;
mso-ascii-font-family:Calibri;
mso-ascii-theme-font:minor-latin;
mso-fareast-font-family:Calibri;
mso-fareast-theme-font:minor-latin;
mso-hansi-font-family:Calibri;
mso-hansi-theme-font:minor-latin;
mso-bidi-font-family:"Times New Roman";
mso-bidi-theme-font:minor-bidi;
mso-font-kerning:0pt;
mso-ligatures:none;}div.WordSection1
{page:WordSection1;}
Friday, August 25, 2023
Buffalo slaughter left lasting impact on Indigenous peoples
Wednesday, August 23, 2023
Biologist gets the scoop on squash bug poop
By Carol Clark
The squash bug carries a gut bacterium that is essential for the bug’s development into an adult. But when they hatch from their eggs, squash bug nymphs do not have the bacteria in their systems. That left scientists who study the interplay between insects and their internal microbes wondering: How do the nymphs acquire these essential microbes?
Jason Chen, an Emory University graduate student in the Department of Biology, stumbled upon a clue one evening in the lab.
He had finished up experiments on some adult squash bugs whose Caballeronia bacteria he had tagged with a red fluorescent protein. The bugs were housed in a plastic box with pieces of paper towel inside as bedding. He tossed some nymphs inside the container just as a place to hold them while he cleaned up for the day.
“When I came back to turn the lights out, I noticed that all the nymphs had flocked around one of the poop spots left on a paper towel by the adults,” Chen says. “Normally nymphs wander around a lot but they had all stopped around this poop. They were transfixed by it. I wondered what that behavior meant.”
He eventually checked the nymphs under a microscope and saw that their guts lit up with the same red fluorescence as the adults. More experiments confirmed the finding — nymph squash bugs eat the feces of adults to acquire the bacteria they need to grow.
Current Biology published the discovery, which may offer insights for improved methods to control the squash bug, a significant agricultural pest.
Read more about the discovery here.
Related:
Thursday, August 10, 2023
Images of enzyme in action reveal secrets of antibiotic-resistant bacteria
By Carol Clark
Bacteria draw from an arsenal of weapons to combat the drugs intended to kill them. Among the most prevalent of these weapons are ribosome-modifying enzymes. These enzymes are growing increasingly common, appearing worldwide in clinical samples in a range of drug-resistant bacteria.
Now scientists have captured the first images of one important class of these enzymes in action. The images show how the enzymes latch onto a particular site on the bacterial ribosome and squeeze it like a pair of tweezers to extract an RNA nucleotide and alter it.
The Proceedings of the National Academy of Sciences (PNAS) published the findings, led by scientists at Emory University. The advanced technique of cryoelectron microscopy made the ultra-high-resolution, three-dimensional snapshots possible.
“Seeing is believing,” says Christine Dunham, Emory professor of chemistry and co-corresponding author of the paper. “The minute you see biological structures interacting in real life at the atomic level it’s like solving a jigsaw puzzle. You see how everything fits together and you get a clearer idea of how things work.”
The insights may lead to the design of new antibiotic therapies to inhibit the drug-resistance activities of RNA methyltransferase enzymes. These enzymes transfer a small hydrocarbon known as a methyl group from one molecule to another, a process known as methylation.
“Methylation is one of the smallest chemical modifications in biology,” says Graeme Conn, professor of biochemistry in Emory’s School of Medicine and co-corresponding author of the paper. “But this tiny modification can fundamentally change biology. In this case, it confers resistance that allows bacteria to evade an entire class of antibiotics.”
Both Conn and Dunham are also members of the Emory Antibiotic Resistance Center.
First author of the paper is Pooja Srinivas, who did the work as a PhD candidate in Emory’s graduate program in molecular and systems pharmacology. She has since graduated and is now a postdoctoral fellow at the University of Washington.
Understanding the ribosome
Dunham is a leading expert on the ribosome — an elaborate structure that operates like a factory within a cell to manufacture proteins. Proteins are the machines that make cells run while nucleic acids such as DNA and RNA store the blueprints for life. The ribosome is made mostly of RNA, which does not just store information but can also act as an enzyme, catalyzing chemical reactions.
One goal of Dunham’s lab is to find ways to manipulate bacterial ribosomes to make them more susceptible to antimicrobials. If an antimicrobial successfully inactivates bacterial ribosomes, that shuts down the manufacturing of proteins essential for bacterial growth and survival.
The idea is to exploit differences in human cellular ribosomes and bacterial ribosomes, so that only the bacteria is targeted by an antimicrobial drug.
Antimicrobials, however, need to get past bacterial defenses.
“It’s like a molecular arms race,” Dunham explains. Bacteria keep evolving new weapons as a defense against drugs, while scientists evolve new strategies to disarm bacteria.
Enzymes that modify the ribosome
Conn is a leading expert in the bacterial defense weapons known as ribosomal RNA methyltransferase enzymes. This family of enzymes was originally discovered in soil bacteria. They are now increasingly found in bacterial infections in people and animals, making these infections harder to treat.
“They keep turning up more and more often in clinical samples of some nasty bacterial pathogens in different parts of the world,” Conn says.
The enzymes can drive deadly drug-resistance in pathogens such as E. coli, Salmonella, Klebsiella pneumoniae, Pseudomonas aeruginosa and Enterobacteriaceae. The enzymes add a methyl group at a specific site on the bacterial ribosome. That addition blocks the ability of a class of antibiotics known as aminoglycosides to bind and cause their antibacterial action.
For the PNAS paper, the researchers focused on a culprit within this family of enzymes known as ribosomal RNA methyltransferase C, or RmtC.
A complicated enzyme
For decades, researchers have relied on a technique known as X-ray crystallography to reveal the atomic details of how molecular machines work when the molecules are arranged in a crystal.
In 2015, for example, Dunham’s lab obtained precise pictures through X-ray crystallography of how an enzyme known as HigB rips up RNA to inhibit growth of the bacteria. By restraining the growth of the bacteria that makes it, HigB establishes a dormant “persister cell” state that makes the bacteria tolerant to antibiotics.
The secrets of how the RmtC enzyme interacts with the ribosome, however, eluded X-ray crystallography.
“RmtC is much more complicated,” Dunham explains. “It’s an interesting enzyme from a basic science perspective because it looks so different from others.”
A resolution revolution
Recent advances in cryoelectron microscopy opened the door to zooming in on the complex mechanisms of RmtC.
Cryoelectron microscopy does not require crystallization to reveal the structures of molecules and how they interact. Instead, liquid samples are frozen rapidly to form a glassy matrix. The glassy matrix retains the three-dimensional structure of molecules and protects them from deterioration by the intense electron beam.
Meisam Nosrati, a former postdoctoral fellow in the Conn lab and a co-author of the PNAS paper, prepared samples of RmtC interacting with part of an E. coli ribosome. He tapped the expertise of co-author Lindsay Comstock, a chemist at Wake Forest University who developed a technique to trap and stabilize the enzyme in the needed position.
Nosrati then froze the samples on a tiny grid and sent them to the Pacific Northwest Center for Cryo-EM for imaging.
As a graduate student in the Dunham lab, Pooja Srinivas then analyzed and interpreted the microscopy dataset. She used computer algorithms to stitch together thousands of individual images.
The result turned the images into a flipbook that revealed the complicated structure of RmtC in action.
“The enzyme latches on like a pincer to the ribosome,” Dunham explains. “It tightens its grip until it squeezes out a nucleotide from the interior of an RNA helix. It then chemically modifies that nucleotide.”
The enzyme is exquisitely specific about where it binds to the ribosome, a huge macromolecule made up of 50 different proteins and 6,000 different RNA nucleotides.
The researchers used biochemistry techniques to validate that what they observed matched previous findings for how RmtC makes bacteria resistant to aminoglycoside antimicrobials that target the ribosome.
Strategies for new therapies
The researchers are now trying to develop new ways to counter the effects of RmtC and related enzymes based on the new information.
“Knowledge of the shape of the enzyme as its performs its chemical reaction gives us new targets to inhibit its effects,” Conn says. “For instance, we could target the pincer action of the enzyme to try to prevent it from squeezing and binding to the ribosome. We now know that the enzyme forms a pocket on its surface where a small molecule might sit to block this action.”
Additional co-authors of the PNAS paper are Natalia Zelinskaya and Debayan Dey, research scientists in the Conn lab. Funding for the work was provided by the National Institutes of Health and the Burroughs Wellcome Fund Investigator in the Pathogenesis of Infectious Disease Award.
Related:
Biochemist Dunham shifts the frame on proteins
New molecule found in chestnut trees disarms dangerous staph bacteria
Monday, August 7, 2023
Physicists open new path to exotic form of superconductivity
By Carol Clark
Physicists have identified a mechanism for the formation of oscillating superconductivity known as pair-density waves. Physical Review Letters published the discovery, which provides new insight into an unconventional superconductive state seen in certain materials, including high-temperature superconductors.
“We discovered that structures known as Van Hove singularities can produce modulating, oscillating states of superconductivity,” says Luiz Santos, assistant professor of physics at Emory University and senior author of the study. “Our work provides a new theoretical framework for understanding the emergence of this behavior, a phenomenon that is not well understood.”
First author of the study is Pedro Castro, an Emory physics graduate student. Co-authors include Daniel Shaffer, a postdoctoral fellow in the Santos group, and Yi-Ming Wu from Stanford University.
The work was funded by the U.S. Department of Energy’s Office of Basic Energy Sciences.
The puzzle of superconductivity
Santos is a theorist who specializes in condensed matter physics. He studies the interactions of quantum materials — tiny things such as atoms, photons and electrons — that don’t behave according to the laws of classical physics.
Superconductivity, or the ability of certain materials to conduct electricity without energy loss when cooled to a super-low temperature, is one example of intriguing quantum behavior. The phenomenon was discovered in 1911 when Dutch physicist Heike Kamerlingh Onnes showed that mercury lost its electrical resistance when cooled to 4 Kelvin or minus 371 degrees Fahrenheit. That’s about the temperature of Uranus, the coldest planet in the solar system.
It took scientists until 1957 to come up with an explanation for how and why superconductivity occurs. At normal temperatures, electrons roam more or less independently. They bump into other particles, causing them to shift speed and direction and dissipate energy. At low temperatures, however, electrons can organize into a new state of matter.
“They form pairs that are bound together into a collective state that behaves like a single entity,” Santos explains. “You can think of them like soldiers in an army. If they are moving in isolation they are easier to deflect. But when they are marching together in lockstep it’s much harder to destabilize them. This collective state carries current in a robust way.”
A holy grail of physics
Superconductivity holds huge potential. In theory, it could allow for electric current to move through wires without heating them up, or losing energy. These wires could then carry far more electricity, far more efficiently.
“One of the holy grails of physics is room-temperature superconductivity that is practical enough for everyday-living applications,” Santos says. “That breakthrough could change the shape of civilization.”
Many physicists and engineers are working on this frontline to revolutionize how electricity gets transferred.
Meanwhile, superconductivity has already found applications. Superconducting coils power electromagnets used in magnetic resonance imaging (MRI) machines for medical diagnostics. A handful of magnetic levitation trains are now operating in the world, built on superconducting magnets that are 10 times stronger than ordinary electromagnets. The magnets repel each other when the matching poles face each other, generating a magnetic field capable of levitating and propelling a train.
The Large Hadron Collider, a particle accelerator that scientists are using to research the fundamental structure of the universe, is another example of technology that runs through superconductivity.
Superconductivity continues to be discovered in more materials, including many that are superconductive at higher temperatures.
An accidental discovery
One focus of Santos’ research is how interactions between electrons can lead to forms of superconductivity that cannot be explained by the 1957 description of superconductivity. An example of this so-called exotic phenomenon is oscillating superconductivity, when the paired electrons dance in waves, changing amplitude.
In an unrelated project, Santos asked Castro to investigate specific properties of Van Hove singularities, structures where many electronic states become close in energy. Castro’s project revealed that the singularities appeared to have the right kind of physics to seed oscillating superconductivity.
That sparked Santos and his collaborators to delve deeper. They uncovered a mechanism that would allow these dancing-wave states of superconductivity to arise from Van Hove singularities.
“As theoretical physicists, we want to be able to predict and classify behavior to understand how nature works,” Santos says. “Then we can start to ask questions with technological relevance.”
Some high-temperature superconductors — which function at temperatures about three times as cold as a household freezer — have this dancing-wave behavior.
The discovery of how this behavior can emerge from Van Hove singularities provides a foundation for experimentalists to explore the realm of possibilities it presents.
“I doubt that Kamerlingh Onnes was thinking about levitation or particle accelerators when he discovered superconductivity,” Santos says. “But everything we learn about the world has potential applications.”
Related: